library(shiny)
library(forcats)
library(tidyverse)
library(here)
library(hablar)
library(janitor)
library(gt)Appendix G — Imaging CRF Data Quality Checks
This document goes over quality checks for the Imaging Case Report Form (CRF). The headings in the sidebar help the user navigate to their desired content.
G.1 Read in Data and write functions
G.1.1 Load Libraries
G.1.2 Function
Write a function to remove columns where all rows have NA, this is will remove duplicate columns for the the Thoracotomy/TKA cohort
not_all_na <- function(x) any(!is.na(x))G.2 CRF Quality checks
G.2.1 Imaging Form
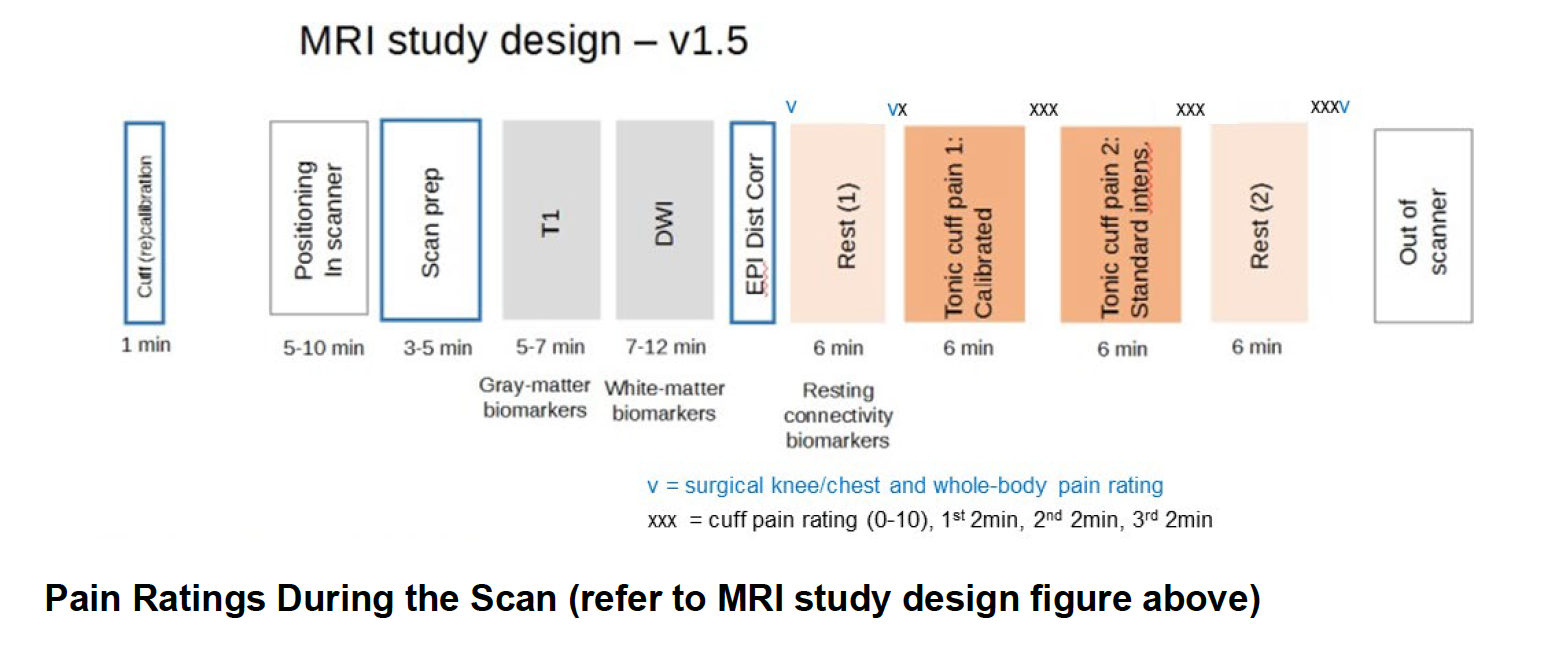
A2CPS: Site Manual of Procedures v3.0 (pg 207)
Read in Imaging data, We will call this img
img <- read_csv(here("data", "imaging", "imaging-2024-11-06.csv"))We will read in QST data since the information on cuff contraindication is recorded in the QST data. We will later merge field names related to cuff contraindication with the imaging data.
# read_csv function results in parsing errors, we will use read.csv instead
tka_img_qst <- read.csv(here("data", "QST", "qst-2024-11-06.csv")) %>%
filter(redcap_repeat_instrument == "qst_mcc1_v03") %>%
select(record_id, redcap_event_name, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, fmricuffcontrayn, fmricuffcalfpressure, fmricuffcalfpressure2, qst_mcc1_v03_complete) %>%
filter(qst_mcc1_v03_complete == 2 & !is.na(redcap_repeat_instance)) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))We will also read in “mask” data and call this “mask.” This data contains information on whether the baseline visit occurred before or after the question related to mask use was implemented. Where “yes” indicates that the baseline visit occurred after the question related to mask use was implemented and “no” indicates that the baseline visit occurred before the question related to mask use was implemented.
mask <- read_csv(here("data", "imaging", "mask.csv"))Remove test records
test_records <- c("10000", "15000", "20000", "25000", "40000", "50000", "60000", "70000", "80000", "90000", "100000", "110000", "120000")
img <- img %>%
filter(!record_id %in% test_records)Create a column for cohort type called “cohort”
img <- img %>%
mutate(cohort = case_when(record_id >= 10000 & record_id < 15000 | record_id >= 25000 ~ "TKA", TRUE ~ "Thoracic"))G.2.2 Data Dictionary
Read in data dictionary and remove duplicate field names
img_dict <- read_csv(here("data", "imaging", "imaging-Data-Dictionary-2024-11-06.csv")) %>% distinct(field_name, .keep_all = TRUE)G.2.3 New field name(s):
Add the field name “cohort” and “after_mask” to the data dictionary
# Create field names
cohort_new_row <- data.frame(
field_name = "cohort",
field_type = "Character",
field_note = "Type of surgical cohort",
select_choices_or_calculations = "TKA,Thoracic"
)
# Create field names
mask_new_row <- data.frame(
field_name = "after_mask",
field_type = "Character",
field_note = "yes indicates that the baseline visit occurred after the question related to mask use was implemented and no indicates that the baseline visit occurred before the question related to mask use was implemented",
select_choices_or_calculations = "yes,no"
)
# Add the new row after the last row
img_dict <- img_dict %>%
slice(1:nrow(.)) %>%
add_row(.after = nrow(.), !!!cohort_new_row)
img_dict <- img_dict %>%
slice(1:nrow(.)) %>%
add_row(.after = nrow(.), !!!mask_new_row)The redcap_repeat_instance should be the same for the Imaging session and Imaging Items form. ML and BA confirmed that the following record_ids have a redcap_repeat_instance mismatch at baseline.
TKA

Thoracic

Proposed solution for data release 2.0: For the IDs above, we compared the time stamps of the Imaging session CRF(s) and the Imaging Items form(s), and retained the Imaging items form(s) with the time stamp matching or close to the Imaging session CRF(s) dates.
img <- img %>%
filter(!(record_id == 10747 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 10796 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25068 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25079 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25080 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25090 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25122 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2 | redcap_repeat_instance == 3)) %>%
filter(!(record_id == 25161 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25171 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 25216 & redcap_repeat_instrument == "imaging_items_v11" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 20226 & redcap_repeat_instrument == "imaging_items_v01" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 20302 & redcap_repeat_instrument == "imaging_items_v01" & redcap_repeat_instance == 2)) %>%
filter(!(record_id == 20144 & redcap_repeat_instrument == "imaging_items_v01" & redcap_repeat_instance == 2))G.2.4 TKA Cohort Imaging
tka_img <- img %>%
select(record_id, guid, redcap_data_access_group, redcap_event_name, redcap_repeat_instrument, redcap_repeat_instance, starts_with("fmri"), imaging_mcc1_v09_complete, cohort)keep subjects from the TKA cohort, with the most recent baseline visit and merge with QST data.
tka_img <- tka_img %>%
filter(cohort == "TKA") %>%
filter(redcap_repeat_instrument == "imaging_mcc1_v09") %>%
filter(imaging_mcc1_v09_complete == 2 & !is.na(redcap_repeat_instance)) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))
tka_img <- left_join(tka_img, tka_img_qst, by = intersect(names(tka_img), names(tka_img_qst)))keep subjects marked as “Yes, all scans were completed” or “Yes, but only the scans indicated below were completed”
irdf.complete <- tka_img %>%
filter(fmricuffcompletescl != 0 & imaging_mcc1_v09_complete == 2)G.2.4.0.1 Flag 1:
In subjects with cuff not contraindicated in the QST form, check if the cuff site was entered for subjects who underwent fMRI individualized pressure and/or fMRI standard pressure.
irerror1 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | fmricuffcpyn == 1 | fmricuffipyn == 1) %>%
filter(is.na(fmricuffleg) & fmricuffcontrayn != 1) %>%
add_column(error_type = "In subjects with cuff not contraindicated in the QST form, check if the cuff site was entered for subjects who underwent fMRI individualized pressure and/or fMRI standard pressure") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.2 Flag 2:
If some scans were completed, check if the type of completed scans were specified
irerror2 <- irdf.complete %>%
filter(fmricuffcompletescl == 2 & (is.na(fmricufft1yn) | is.na(fmricuffdwiyn) | is.na(fmricuffrest1yn) |
is.na(fmricuffipyn) | is.na(fmricuffcpyn) | is.na(fmricuffrest2yn))) %>%
add_column(error_type = "some scans were completed but type was not specified") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.3 Flag 3:
In subjects marked as “Yes, all scans were completed” OR “Yes, but only the scans indicated below were completed” AND cuff was applied AND recalibration of pressure was needed, check for mismatch between recalibration pressures “fmricuffcalfpressurerecal” and “fmricuffcalfpressurerecal2” (pressure 2 double entry) or missing values.
irerror3 <- irdf.complete %>%
filter(!is.na(fmricuffleg)) %>%
filter(fmricuffcontrarecal == 1) %>%
mutate(pressure_diff = fmricuffcalfpressurerecal - fmricuffcalfpressurerecal2) %>%
filter(pressure_diff != 0 | is.na(pressure_diff)) %>%
select(record_id, redcap_event_name, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, fmricuffcompletescl, redcap_event_name, fmricuffcontrarecal, fmricuffleg, fmricuffcalfpressurerecal, fmricuffcalfpressurerecal2) %>%
add_column(error_type = "Mismatch between recalibration pressures or missing values") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.4 Flag 4:
T1 Scan:
In subjects with T1 scanning, check if the quality of the T1 scan was rated on the first scan or the repeated scan.
irerror4 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricufft1yn) & fmricufft1yn == 1)) %>%
filter(is.na(fmricufft1techrating) | fmricufft1repeated == 1 & is.na(fmricufft1techrating2)) %>%
select(record_id, redcap_event_name, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, fmricuffcompletescl, redcap_event_name, fmricuffcontrarecal, fmricuffleg, fmricuffcalfpressurerecal, fmricuffcalfpressurerecal2) %>%
add_column(error_type = "T1 scan quality not rated") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.5 Flag 5:
Before first resting state:
In subjects with resting-state scan performed, check if there were missing values for surgical site pain or if there was a mismatch between entries for surgical site pain.
irerror5 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(surg.pain.diff1 = fmricuffrestpainss - fmricuffrestpainss2) %>%
filter(surg.pain.diff1 != 0 | is.na(surg.pain.diff1)) %>%
as_tibble() %>%
add_column(error_type = "Before resting-state scan performed: Check if there were missing values for surgical site pain or if there was a mismatch between entries for surgical site pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.6 Flag 6:
Before first resting state:
In subjects with resting-state scan performed, check if there were missing values for overall body pain or if there was a mismatch between entries for overall body pain.
irerror6 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(all.pain.diff1 = fmricuffrestpainovrall - fmricuffrestpainovrall2) %>%
filter(all.pain.diff1 != 0 | is.na(all.pain.diff1)) %>%
as_tibble() %>%
add_column(error_type = "Before resting-state scan performed, check if there were missing values for overall body pain or was a mismatch between entries for overall body pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.7 Flag 7:
After first resting fMRI scan:
In subjects with resting-state scan performed, check if there were missing values for surgical site pain or was a mismatch between entries for surgical site pain.
irerror7 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(surg.pain.diff2 = fmricuffcurrpainaftfirstscanss - fmricuffcurrpainaftfirstscanss2) %>%
filter(surg.pain.diff2 != 0 | is.na(surg.pain.diff2)) %>%
as_tibble() %>%
add_column(error_type = "After first resting-state scan performed, check if there were missing values for surgical site pain or was a mismatch between entries for surgical site pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.8 Flag 8:
After first resting fMRI scan:
Check if there were missing values for overall body pain or if there was a mismatch between entries for overall body pain.
irerror8 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(all.pain.diff2 = fmricuffcurrpainaftfirstscanovrall - fmricuffcurrpainaftfirstscanovrall2) %>%
filter(all.pain.diff2 != 0 | is.na(all.pain.diff2)) %>%
as_tibble() %>%
add_column(error_type = "After resting-state scan performed, check if there were missing values for overall body pain or if there was a mismatch between entries for overall body pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.9 Flag 9:
After first resting fMRI scan:
Check if baseline cuff pain was missing in subjects with fMRI individualized and standard pressures performed (if standard and personalized/individualized were not performed, it is assumed that cuff was C/I)
irerror9 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
filter(fmricuffipyn == 1 & fmricuffcpyn == 1) %>%
mutate(cuff.pain.diff1 = fmricuffcurrpainaftfirstscancp - fmricuffcurrpainaftfirstscancp2) %>%
filter(cuff.pain.diff1 != 0 | is.na(cuff.pain.diff1)) %>%
add_column(error_type = "Check if baseline cuff pain was missing in subjects with fMRI individualized and standard pressures performed") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.10 Flag 10:
After first cuff fMRI scan (personalized cuff pressure):
Check if there are missing values or mismatch between cuff pain at the beginning of the scan.
irerror10 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffipyn) & fmricuffipyn == 1)) %>%
mutate(cuff.begin.diff1 = fmricuffpainbegin - fmricuffpainbegin2) %>%
filter(cuff.begin.diff1 != 0 | is.na(cuff.begin.diff1)) %>%
add_column(error_type = "After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the beginning of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.11 Flag 11:
After first cuff fMRI scan (personalized cuff pressure):
Check if there are missing values or mismatch between cuff pain during mid scan.
irerror11 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffipyn) & fmricuffipyn == 1)) %>%
mutate(cuff.mid.diff1 = fmricuffpainmid - fmricuffpainmid2) %>%
filter(cuff.mid.diff1 != 0 | is.na(cuff.mid.diff1)) %>%
as_tibble() %>%
add_column(error_type = "After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch during mid scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.12 Flag 12:
After first cuff fMRI scan (personalized cuff pressure):
Check if there are missing values or mismatch between cuff pain at the end of the scan.
irerror12 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffipyn) & fmricuffipyn == 1)) %>%
mutate(cuff.end.diff1 = fmricuffpainend - fmricuffpainend2) %>%
filter(cuff.end.diff1 != 0 | is.na(cuff.end.diff1)) %>%
add_column(error_type = "After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the end of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.13 Flag 13:
After second cuff fMRI scan (constant 120 mmHg pressure):
Check if there are missing values or mismatch between cuff pain at the beginning of the scan.
irerror13 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffcpyn) & fmricuffcpyn == 1)) %>%
mutate(cuff.begin.diff2 = fmricuffpaincpbegin - fmricuffpaincpbegin2) %>%
filter(cuff.begin.diff2 != 0 | is.na(cuff.begin.diff2)) %>%
add_column(error_type = "After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the beginning of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.14 Flag 14:
After second cuff fMRI scan (constant 120 mmHg pressure):
Check if there are missing values or mismatch between cuff pain during mid scan.
irerror14 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffcpyn) & fmricuffcpyn == 1)) %>%
mutate(cuff.mid.diff2 = fmricuffpaincpmid - fmricuffpaincpmid2) %>%
filter(cuff.mid.diff2 != 0 | is.na(cuff.mid.diff2)) %>%
add_column(error_type = "After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain during mid scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.15 Flag 15:
After second cuff fMRI scan (constant 120 mmHg pressure):
Check if there are missing values or mismatch between cuff pain at the end of the scan.
irerror15 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffcpyn) & fmricuffcpyn == 1)) %>%
mutate(cuff.end.diff2 = fmricuffpaincpend - fmricuffpaincpend2) %>%
filter(cuff.end.diff2 != 0 | is.na(cuff.end.diff2)) %>%
as_tibble() %>%
add_column(error_type = "After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the end of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.16 Flag 16:
After last resting fMRI scan:
Check if there are missing values or mismatch between residual cuff pain at the beginning of the scan:
irerror16 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffcpyn) & fmricuffcpyn == 1 & fmricuffrest2yn == 1) | (!is.na(fmricuffipyn) & fmricuffipyn == 1 & fmricuffrest2yn == 1)) %>%
mutate(rest.cuff.begin.diff1 = fmricuffpainrestscanbegin - fmricuffpainrestscanbegin2) %>%
filter(rest.cuff.begin.diff1 != 0 | is.na(rest.cuff.begin.diff1)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the beginning of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.17 Flag 17:
After last resting fMRI scan:
Check if there are missing values or mismatch between residual cuff pain during mid scan
irerror17 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffcpyn) & fmricuffcpyn == 1 & fmricuffrest2yn == 1) | (!is.na(fmricuffipyn) & fmricuffipyn == 1 & fmricuffrest2yn == 1)) %>%
mutate(rest.cuff.mid.diff1 = fmricuffpainrestscanmid - fmricuffpainrestscanmid2) %>%
filter(rest.cuff.mid.diff1 != 0 | is.na(rest.cuff.mid.diff1)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain during mid scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.18 Flag 18:
After last resting fMRI scan:
Check if there are missing values or mismatch between residual cuff pain at the end of the scan:
irerror18 <- irdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffcpyn) & fmricuffcpyn == 1 & fmricuffrest2yn == 1) | (!is.na(fmricuffipyn) & fmricuffipyn == 1 & fmricuffrest2yn == 1)) %>%
mutate(rest.cuff.end.diff1 = fmricuffpainrestscanend - fmricuffpainrestscanend2) %>%
filter(rest.cuff.end.diff1 != 0 | is.na(rest.cuff.end.diff1)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the end of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.19 Flag 19:
After last resting fMRI scan:
Check if there are missing values or mismatch between surgical site pain
irerror19 <- irdf.complete %>%
filter((!is.na(fmricuffrest2yn) & fmricuffrest2yn == 1) | fmricuffcompletescl == 1) %>%
mutate(surg.diff3 = fmricuffpainaftlastscanss - fmricuffpainaftlastscanss2) %>%
filter(surg.diff3 != 0 | is.na(surg.diff3)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between surgical site pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.20 Flag 20:
After last resting fMRI scan: Check if there are missing values or mismatch between overall body pain.
irerror20 <- irdf.complete %>%
filter((!is.na(fmricuffrest2yn) & fmricuffrest2yn == 1) | fmricuffcompletescl == 1) %>%
mutate(body.diff3 = fmricuffpainaftlastscanany - fmricuffpainaftlastscanany2) %>%
filter(body.diff3 != 0 | is.na(body.diff3)) %>%
add_column(error_type = "After last resting fMRI scan:Check if there are missing values or mismatch between overall body pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.21 Flag 21:
If a visit is marked complete and DICOM files were uploaded, check if “Test Completed” (fmricuffcompletescl) is missing.
irerror21 <- tka_img %>%
filter(imaging_mcc1_v09_complete == 2) %>%
filter(is.na(fmricuffcompletescl)) %>%
filter(fmricuffdicuploaded == 1) %>%
as_tibble() %>%
add_column(error_type = "IF a visit is marked complete and DICOM files were uploaded, check if Test Completed (fmricuffcompletescl) is missing") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.22 Flag 22
Check if any of the scans were completed but information on “mask worn” was missing. We will first merge with the “mask” data to account for visits that occurred before the question related to make use was implemented.
tka_img <- left_join(tka_img, mask, by = intersect(names(tka_img), names(mask)))
irerror22 <- tka_img %>%
filter(imaging_mcc1_v09_complete == 2) %>%
filter(fmricuffcompletescl == 1 | fmricufft1yn == 1 | fmricuffdwiyn == 1 | fmricuffrest1yn == 1 |
fmricuffipyn == 1 | fmricuffcpyn == 1 | fmricuffrest2yn == 1) %>%
filter(is.na(fmri_face_mask)) %>%
add_column(error_type = "Check if any of the scans were completed but information on mask worn was missing") %>%
add_column(errors = "error") %>%
filter(after_mask == "yes") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.4.0.23 Create Imaging error report for the TKA cohort.
# Specify common prefix
m1img_error <- "irerror"
# Find data frames in the global environment with the specified prefix
m1img_list <- mget(ls(pattern = paste0("^", m1img_error)))
# Combine the data frames using bind_rows
m1img_report <- bind_rows(m1img_list) %>%
pivot_wider(names_from = "error_type", values_from = "errors") %>%
mutate_all(~ replace_na(., ""))m1img_report %>%
gt() %>%
tab_header(
title = md("**TKA Imaging Error Report**")
) %>%
tab_options(
table.font.size = px(12),
column_labels.font.size = px(12)
) %>%
tab_style(
style = list(cell_fill(color = "#F4F4F4")),
locations = cells_body(columns = record_id)
)| TKA Imaging Error Report | ||||||||||||||||||||
| record_id | redcap_data_access_group | redcap_repeat_instrument | redcap_repeat_instance | After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the beginning of the scan | After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch during mid scan | After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the end of the scan | After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the beginning of the scan | After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain during mid scan | After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the end of the scan | After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the beginning of the scan | After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain during mid scan | After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the end of the scan | After last resting fMRI scan: Check if there are missing values or mismatch between surgical site pain | After last resting fMRI scan:Check if there are missing values or mismatch between overall body pain | Check if any of the scans were completed but information on mask worn was missing | T1 scan quality not rated | Before resting-state scan performed: Check if there were missing values for surgical site pain or if there was a mismatch between entries for surgical site pain | Before resting-state scan performed, check if there were missing values for overall body pain or was a mismatch between entries for overall body pain | After first resting-state scan performed, check if there were missing values for surgical site pain or was a mismatch between entries for surgical site pain | After resting-state scan performed, check if there were missing values for overall body pain or if there was a mismatch between entries for overall body pain |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10135 | uchicago | imaging_mcc1_v09 | 1 | error | error | error | error | error | error | error | error | |||||||||
| 10506 | uchicago | imaging_mcc1_v09 | 1 | error | error | error | ||||||||||||||
| 25048 | university_of_mich | imaging_mcc1_v09 | 1 | error | error | error | ||||||||||||||
| 10182 | northshore | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10428 | northshore | imaging_mcc1_v09 | 1 | error | error | error | error | |||||||||||||
| 10789 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10095 | northshore | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10513 | uchicago | imaging_mcc1_v09 | 1 | error | error | error | ||||||||||||||
| 10732 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10350 | uchicago | imaging_mcc1_v09 | 1 | error | error | error | error | error | ||||||||||||
| 25029 | university_of_mich | imaging_mcc1_v09 | 1 | error | error | error | error | error | ||||||||||||
| 10416 | uchicago | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10809 | rush_university_me | imaging_mcc1_v09 | 1 | error | error | error | error | error | error | |||||||||||
| 25009 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 25059 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 25068 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 25099 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 25120 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10067 | uchicago | imaging_mcc1_v09 | 1 | error | error | error | ||||||||||||||
| 10235 | uchicago | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10237 | uchicago | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10282 | rush_university_me | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10423 | uchicago | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10706 | uchicago | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10591 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10598 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10610 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 25039 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
| 10040 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10335 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 10603 | uchicago | imaging_mcc1_v09 | 1 | error | error | |||||||||||||||
| 25046 | university_of_mich | imaging_mcc1_v09 | 1 | error | ||||||||||||||||
G.2.4.0.24 Save:
Save “tka_img” and updated data dictionary as .csv filea in the folder named “reformatted_imaging”
write_csv(tka_img, file = here::here("data", "imaging", "Reformatted", "reformatted_tka_img.csv"))
write_csv(img_dict, file = here::here("data", "imaging", "Reformatted", "updated_img_dict.csv"))G.2.5 Thoracotomy Cohort Imaging
thor_img <- img %>%
select(record_id, guid, redcap_data_access_group, redcap_event_name, redcap_repeat_instrument, redcap_repeat_instance, starts_with("fmri"), imaging_mcc2_v01_complete, cohort)
# read_csv function results in parsing errors, we will use read.csv instead
thor_img_qst <- read.csv(here("data", "QST", "qst-2024-11-06.csv")) %>%
filter(redcap_repeat_instrument == "qst_mcc1_v03" & !is.na(redcap_repeat_instance)) %>%
select(record_id, redcap_event_name, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, cuffpfmricontrainddomyn, cuffpfmricontraindyn, fmricuffcalfpressure, fmricuffcalfpressure2, qst_mcc1_v03_complete) %>%
filter(qst_mcc1_v03_complete == 2) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))keep subjects from the Thoracotomy cohort, with the most recent baseline visit and merge with QST data
thor_img <- thor_img %>%
filter(cohort == "Thoracic") %>%
filter(redcap_repeat_instrument == "imaging_mcc2_v01") %>%
filter(imaging_mcc2_v01_complete == 2 & !is.na(redcap_repeat_instance)) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))
thor_img <- left_join(thor_img, thor_img_qst, by = intersect(names(thor_img), names(thor_img_qst)))keep subjects marked as “Yes, all scans were completed” or “Yes, but only the scans indicated below were completed” for fmricuffcompletescl.
tirdf.complete <- thor_img %>%
filter(fmricuffcompletescl != 0 & imaging_mcc2_v01_complete == 2)G.2.5.0.1 Flag 1:
In subjects with cuff not contraindicated in the QST form, check if cuff site was entered for subjects who underwent fMRI individualized pressure and/or fMRI standard pressure.
tirerror1 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | fmricuffspyn == 1 | fmricuffipyn == 1) %>%
filter(is.na(fmricuffleg) & (cuffpfmricontrainddomyn != 1 | cuffpfmricontraindyn != 1)) %>%
add_column(error_type = "In subjects with cuff not contraindicated in the QST form, check if cuff site was entered for subjects who underwent fMRI individualized pressure and/or fMRI standard pressure") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.2 Flag 2:
If some scans were completed, check if the type of completed scans were specified
tirerror2 <- tirdf.complete %>%
filter(fmricuffcompletescl == 2 & (is.na(fmricufft1yn) | is.na(fmricuffdwiyn) | is.na(fmricuffrest1yn) | is.na(fmricuffipyn) | is.na(fmricuffspyn) | is.na(fmricuffrest2yn))) %>%
add_column(error_type = "some scans were completed but type was not specified") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.3 Flag 3:
In subjects marked as “Yes, all scans were completed” OR “Yes, but only the scans indicated below were completed” AND cuff was applied AND recalibration of pressure was needed, check for mismatch between recalibration pressures “fmricuffcalfpressurerecal” and “fmricuffcalfpressurerecal2” (pressure 2 double entry) or missing values.
tirerror3 <- tirdf.complete %>%
filter(!is.na(fmricuffleg)) %>%
filter(fmricuffcontrarecal == 1) %>%
mutate(pressure_diff = fmricuffcalfpressurerecal - fmricuffcalfpressurerecal2) %>%
filter(pressure_diff != 0 | is.na(pressure_diff)) %>%
select(record_id, redcap_event_name, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, fmricuffcompletescl, redcap_event_name, fmricuffcontrarecal, fmricuffleg, fmricuffcalfpressurerecal, fmricuffcalfpressurerecal2) %>%
add_column(error_type = "Mismatch between recalibration pressures or missing values") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.4 Flag 4:
T1 Scan:
In subjects with T1 scanning, check if the quality of the T1 scan was rated on the first scan or the repeated scan.
tirerror4 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricufft1yn) & fmricufft1yn == 1)) %>%
filter(is.na(fmricufft1techrating) | fmricufft1repeated == 1 & is.na(fmricufft1techrating2)) %>%
select(record_id, redcap_event_name, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, fmricuffcompletescl, redcap_event_name, fmricuffcontrarecal, fmricuffleg, fmricuffcalfpressurerecal, fmricuffcalfpressurerecal2) %>%
add_column(error_type = "T1 scan quality not rated") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.5 Flag 5:
Before first resting state:
In subjects with resting-state scan performed, check if there were missing values for surgical site pain or if there was a mismatch between entries for surgical site pain.
tirerror5 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(surg.pain.diff1 = fmricuffrestpainss - fmricuffrestpainss2) %>%
filter(surg.pain.diff1 != 0 | is.na(surg.pain.diff1)) %>%
as_tibble() %>%
add_column(error_type = "Before resting-state scan performed: Check if there were missing values for surgical site pain or if there was a mismatch between entries for surgical site pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.6 Flag 6:
Before first resting state:
In subjects with resting-state scan performed, check if there were missing values for overall body pain or if there was a mismatch between entries for overall body pain.
tirerror6 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(all.pain.diff1 = fmricuffrestpainovrall - fmricuffrestpainovrall2) %>%
filter(all.pain.diff1 != 0 | is.na(all.pain.diff1)) %>%
as_tibble() %>%
add_column(error_type = "Before resting-state scan performed, check if there were missing values for overall body pain or was a mismatch between entries for overall body pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.7 Flag 7:
After first resting fMRI scan:
In subjects with resting-state scan performed, check if there were missing values for surgical site pain or was a mismatch between entries for surgical site pain.
tirerror7 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(surg.pain.diff2 = fmricuffcurrpainaftfirstscanss - fmricuffcurrpainaftfirstscanss2) %>%
filter(surg.pain.diff2 != 0 | is.na(surg.pain.diff2)) %>%
as_tibble() %>%
add_column(error_type = "After first resting-state scan performed, check if there were missing values for surgical site pain or was a mismatch between entries for surgical site pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.8 Flag 8:
After first resting fMRI scan:
Check if there were missing values for overall body pain or if there was a mismatch between entries for overall body pain.
tirerror8 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
mutate(all.pain.diff2 = fmricuffcurrpainaftfirstscanovrall - fmricuffcurrpainaftfirstscanovrall2) %>%
filter(all.pain.diff2 != 0 | is.na(all.pain.diff2)) %>%
as_tibble() %>%
add_column(error_type = "After resting-state scan performed, check if there were missing values for overall body pain or if was a mismatch between entries for overall body pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.9 Flag 9:
After first resting fMRI scan:
Check if baseline cuff pain was missing in subjects with fMRI individualized and standard pressures performed (if standard and personalized/individualized were not performed, it is assumed that cuff was C/I)
tirerror9 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffrest1yn) & fmricuffrest1yn == 1)) %>%
filter(fmricuffipyn == 1 & fmricuffspyn == 1) %>%
mutate(cuff.pain.diff1 = fmricuffcurrpainaftfirstscancuff - fmricuffcurrpainaftfirstscancuff2) %>%
filter(cuff.pain.diff1 != 0 | is.na(cuff.pain.diff1)) %>%
add_column(error_type = "Check if baseline cuff pain was missing in subjects with fMRI individualized and standard pressures performed") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.10 Flag 10:
After first cuff fMRI scan (personalized cuff pressure):
Check if there are missing values or mismatch between cuff pain at the beginning of the scan.
tirerror10 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffipyn) & fmricuffipyn == 1)) %>%
mutate(cuff.begin.diff1 = fmricuffpainbegin - fmricuffpainbegin2) %>%
filter(cuff.begin.diff1 != 0 | is.na(cuff.begin.diff1)) %>%
add_column(error_type = "After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the beginning of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.11 Flag 11:
After first cuff fMRI scan (personalized cuff pressure):
Check if there are missing values or mismatch between cuff pain during mid scan.
tirerror11 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (fmricuffcompletescl == 2 & !is.na(fmricuffipyn) & fmricuffipyn == 1)) %>%
mutate(cuff.mid.diff1 = fmricuffpainmid - fmricuffpainmid2) %>%
filter(cuff.mid.diff1 != 0 | is.na(cuff.mid.diff1)) %>%
as_tibble() %>%
add_column(error_type = "After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch during mid scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.12 Flag 12:
After first cuff fMRI scan (personalized cuff pressure):
Check if there are missing values or mismatch between cuff pain at the end of the scan.
tirerror12 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffipyn) & fmricuffipyn == 1)) %>%
mutate(cuff.end.diff1 = fmricuffpainend - fmricuffpainend2) %>%
filter(cuff.end.diff1 != 0 | is.na(cuff.end.diff1)) %>%
add_column(error_type = "After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the end of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.13 Flag 13:
After second cuff fMRI scan (constant 120 mmHg pressure):
Check if there are missing values or mismatch between cuff pain at the beginning of the scan.
tirerror13 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffspyn) & fmricuffspyn == 1)) %>%
mutate(cuff.begin.diff2 = fmricuffpaincpbegin - fmricuffpaincpbegin2) %>%
filter(cuff.begin.diff2 != 0 | is.na(cuff.begin.diff2)) %>%
add_column(error_type = "After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the beginning of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.14 Flag 14:
After second cuff fMRI scan (constant 120 mmHg pressure):
Check if there are missing values or mismatch between cuff pain during mid scan.
tirerror14 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffspyn) & fmricuffspyn == 1)) %>%
mutate(cuff.mid.diff2 = fmricuffpaincpmid - fmricuffpaincpmid2) %>%
filter(cuff.mid.diff2 != 0 | is.na(cuff.mid.diff2)) %>%
add_column(error_type = "After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain during mid scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.15 Flag 15:
After second cuff fMRI scan (constant 120 mmHg pressure):
Check if there are missing values or mismatch between cuff pain at the end of the scan.
tirerror15 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffspyn) & fmricuffspyn == 1)) %>%
mutate(cuff.end.diff2 = fmricuffpaincpend - fmricuffpaincpend2) %>%
filter(cuff.end.diff2 != 0 | is.na(cuff.end.diff2)) %>%
as_tibble() %>%
add_column(error_type = "After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the end of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.16 Flag 16:
After last resting fMRI scan:
Check if there are missing values or mismatch between residual cuff pain at the beginning of the scan:
tirerror16 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffspyn) & fmricuffspyn == 1 & fmricuffrest2yn == 1) | (!is.na(fmricuffipyn) & fmricuffipyn == 1 & fmricuffrest2yn == 1)) %>%
mutate(rest.cuff.begin.diff1 = fmricuffpainrestscanbegin - fmricuffpainrestscanbegin2) %>%
filter(rest.cuff.begin.diff1 != 0 | is.na(rest.cuff.begin.diff1)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the beginning of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.17 Flag 17:
After last resting fMRI scan:
Check if there are missing values or mismatch between residual cuff pain during mid scan.
tirerror17 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffspyn) & fmricuffspyn == 1 & fmricuffrest2yn == 1) | (!is.na(fmricuffipyn) & fmricuffipyn == 1 & fmricuffrest2yn == 1)) %>%
mutate(rest.cuff.mid.diff1 = fmricuffpainrestscanmid - fmricuffpainrestscanmid2) %>%
filter(rest.cuff.mid.diff1 != 0 | is.na(rest.cuff.mid.diff1)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain during mid scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.18 Flag 18:
After last resting fMRI scan:
Check if there are missing values or mismatch between residual cuff pain at the end of the scan.
tirerror18 <- tirdf.complete %>%
filter(fmricuffcompletescl == 1 | (!is.na(fmricuffspyn) & fmricuffspyn == 1 & fmricuffrest2yn == 1) | (!is.na(fmricuffipyn) & fmricuffipyn == 1 & fmricuffrest2yn == 1)) %>%
mutate(rest.cuff.end.diff1 = fmricuffpainrestscanend - fmricuffpainrestscanend2) %>%
filter(rest.cuff.end.diff1 != 0 | is.na(rest.cuff.end.diff1)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the end of the scan") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.19 Flag 19:
After last resting fMRI scan:
Check if there are missing values or mismatch between surgical site pain
tirerror19 <- tirdf.complete %>%
filter((!is.na(fmricuffrest2yn) & fmricuffrest2yn == 1) | fmricuffcompletescl == 1) %>%
mutate(surg.diff3 = fmricuffpainaftlastscanss - fmricuffpainaftlastscanss2) %>%
filter(surg.diff3 != 0 | is.na(surg.diff3)) %>%
add_column(error_type = "After last resting fMRI scan: Check if there are missing values or mismatch between surgical site pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.20 Flag 20:
After last resting fMRI scan: Check if there are missing values or mismatch between overall body pain.
tirerror20 <- tirdf.complete %>%
filter((!is.na(fmricuffrest2yn) & fmricuffrest2yn == 1) | fmricuffcompletescl == 1) %>%
mutate(body.diff3 = fmricuffpainaftlastscanany - fmricuffpainaftlastscanany2) %>%
filter(body.diff3 != 0 | is.na(body.diff3)) %>%
add_column(error_type = "After last resting fMRI scan:Check if there are missing values or mismatch between overall body pain") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.21 Flag 21:
IF a visit is marked complete and DICOM files were uploaded, check if “Test Completed” (fmricuffcompletescl) is missing.
tirerror21 <- thor_img %>%
filter(imaging_mcc2_v01_complete == 2) %>%
filter(is.na(fmricuffcompletescl)) %>%
filter(fmricuffdicuploaded == 1) %>%
as_tibble() %>%
add_column(error_type = "If a visit is marked complete and DICOM files were uploaded, check if Test Completed (fmricuffcompletescl) is missing") %>%
add_column(errors = "error") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.22 Flag 22
Check if any of the scans were completed but information on “mask worn” was missing. We will first merge with the “mask” data to account for visits that occurred before the question related to make use was implemented.
thor_img <- left_join(thor_img, mask, by = intersect(names(thor_img), names(mask)))
tirerror22 <- thor_img %>%
filter(imaging_mcc2_v01_complete == 2) %>%
filter(fmricuffcompletescl == 1 | fmricufft1yn == 1 | fmricuffdwiyn == 1 | fmricuffrest1yn == 1 |
fmricuffipyn == 1 | fmricuffspyn == 1 | fmricuffrest2yn == 1) %>%
filter(is.na(fmri_face_mask)) %>%
add_column(error_type = "Check if any of the scans were completed but information on mask worn was missing") %>%
add_column(errors = "error") %>%
filter(after_mask == "yes") %>%
select(record_id, redcap_data_access_group, redcap_repeat_instrument, redcap_repeat_instance, error_type, errors)G.2.5.0.23 Create Imaging error report for the Thoracotomy cohort.
# Specify common prefix
m2img_error <- "tirerror"
# Find data frames in the global environment with the specified prefix
m2img_list <- mget(ls(pattern = paste0("^", m2img_error)))
# Combine the data frames using bind_rows
m2img_report <- bind_rows(m2img_list) %>%
pivot_wider(names_from = "error_type", values_from = "errors") %>%
mutate_all(~ replace_na(., ""))m2img_report %>%
gt() %>%
tab_header(
title = md("**Thoracotomy Cohort Imaging Error Report**")
) %>%
tab_options(
table.font.size = px(12),
column_labels.font.size = px(12)
) %>%
tab_style(
style = list(cell_fill(color = "#F4F4F4")),
locations = cells_body(columns = record_id)
)| Thoracotomy Cohort Imaging Error Report | ||||||||||||||||
| record_id | redcap_data_access_group | redcap_repeat_instrument | redcap_repeat_instance | After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch during mid scan | After first cuff fMRI scan (personalized cuff pressure):check if there are missing values or mismatch between cuff pain at the end of the scan | After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the beginning of the scan | After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain during mid scan | After second cuff fMRI scan (constant 120 mmHg pressure):Check if there are missing values or mismatch between cuff pain at the end of the scan | After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the beginning of the scan | After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain during mid scan | After last resting fMRI scan: Check if there are missing values or mismatch between residual cuff pain at the end of the scan | After last resting fMRI scan:Check if there are missing values or mismatch between overall body pain | Check if any of the scans were completed but information on mask worn was missing | T1 scan quality not rated | After first resting-state scan performed, check if there were missing values for surgical site pain or was a mismatch between entries for surgical site pain | After resting-state scan performed, check if there were missing values for overall body pain or if was a mismatch between entries for overall body pain |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20252 | university_of_mich | imaging_mcc2_v01 | 1 | error | error | error | error | |||||||||
| 20018 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20214 | spectrum_health | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20336 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20016 | university_of_mich | imaging_mcc2_v01 | 1 | error | error | error | ||||||||||
| 20046 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20004 | university_of_mich | imaging_mcc2_v01 | 1 | error | error | error | error | |||||||||
| 20128 | university_of_mich | imaging_mcc2_v01 | 1 | error | error | |||||||||||
| 20043 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20049 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20175 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20311 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20317 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20378 | spectrum_health | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20390 | spectrum_health | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20007 | university_of_mich | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20153 | spectrum_health | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20319 | spectrum_health | imaging_mcc2_v01 | 1 | error | ||||||||||||
| 20130 | university_of_mich | imaging_mcc2_v01 | 1 | error | error | |||||||||||
G.2.5.0.24 Save:
Save “thor_img” and data dictionary as .csv files in the folder named “reformatted_imaging”
write_csv(thor_img, file = here::here("data", "imaging", "Reformatted", "reformatted_thor_img.csv"))
write_csv(img_dict, file = here::here("data", "imaging", "Reformatted", "updated_img_dict.csv"))G.2.6 TKA cohort Imaging Items v1.1 form:
tka_img_items <- img %>%
select(record_id, guid, redcap_data_access_group, redcap_event_name, redcap_repeat_instrument, redcap_repeat_instance, starts_with("img"), imaging_items_v11_complete, cohort)keep subjects from the TKA cohort, with the most recent baseline visit.
tka_img_items <- tka_img_items %>%
filter(cohort == "TKA") %>%
filter(redcap_repeat_instrument == "imaging_items_v11") %>%
filter(imaging_items_v11_complete == 2 & !is.na(redcap_repeat_instance)) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))G.2.6.0.1 Save:
Save “tka_img_items” and data dictionary as .csv files in the folder named “Reformatted”
write_csv(tka_img_items, file = here::here("data", "imaging", "Reformatted", "reformatted_tka_img_items.csv"))G.2.7 Thoracotomy cohort Imaging Items v1.1 form: form:
thor_img_items <- img %>%
select(record_id, guid, redcap_data_access_group, redcap_event_name, redcap_repeat_instrument, redcap_repeat_instance, starts_with("img"), imaging_items_v01_complete, cohort)keep subjects from the TKA cohort, with the most recent baseline visit.
thor_img_items <- thor_img_items %>%
filter(cohort == "Thoracic") %>%
filter(redcap_repeat_instrument == "imaging_items_v01") %>%
filter(imaging_items_v01_complete == 2 & !is.na(redcap_repeat_instance)) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))G.2.7.0.1 Save:
Save “thor_meds” and data dictionary as .csv files in the folder named “Reformatted”
write_csv(thor_img_items, file = here::here("data", "imaging", "Reformatted", "reformatted_thor_img_items.csv"))