library(shiny)
library(forcats)
library(tidyverse)
library(here)
library(hablar)
library(janitor)
library(gt)Appendix E — Biospecimen Collection CRF Data Quality Checks
This document goes over quality checks for the Biospecimen Collection Case Report Form (CRF). The headings in the sidebar help the user navigate to their desired content.
E.1 Read in Data and write functions
E.1.1 Load Libraries
E.1.2 Function
Write a function to remove columns where all rows have NA, this is will remove duplicate columns for the the Thoracotomy/TKA cohort
not_all_na <- function(x) any(!is.na(x))E.2 CRF Quality checks
E.2.1 Biospecimen collection Form
Read in Biospecimen collection data, We will call this bio
bio <- read_csv(here("data", "blood-draw", "blood-draw-2024-11-06.csv"))Remove test records
test_records <- c(
"10000",
"15000",
"20000",
"25000",
"40000",
"50000",
"60000",
"70000",
"80000",
"90000",
"100000",
"110000",
"120000"
)
bio <- bio %>%
filter(!record_id %in% test_records)Create a column for cohort type called “cohort”
bio <- bio %>%
mutate(
cohort = case_when(
record_id >= 10000 & record_id < 15000 | record_id >= 25000 ~ "TKA",
TRUE ~ "Thoracic"
)
)E.2.2 Data Dictionary
Read in data dictionary and remove duplicate field names
bio_dict <- read_csv(here(
"data",
"blood-draw",
"blood-draw-Data-Dictionary-2024-11-06.csv"
)) %>%
distinct(field_name, .keep_all = TRUE)E.2.3 New field name(s)
Add the field name “cohort” to the data dictionary
# Create field names
cohort_new_row <- data.frame(
field_name = "cohort",
field_type = "Character",
field_note = "Type of surgical cohort",
select_choices_or_calculations = "TKA,Thoracic"
)
# Add the new row after the last row
bio_dict <- bio_dict %>%
slice(1:nrow(.)) %>%
add_row(.after = nrow(.), !!!cohort_new_row)The redcap_repeat_instance should be the same for the blood draw and current medications form. ML and BA confirmed that the following record_ids have a redcap_repeat_instance mismatch at baseline.
TKA
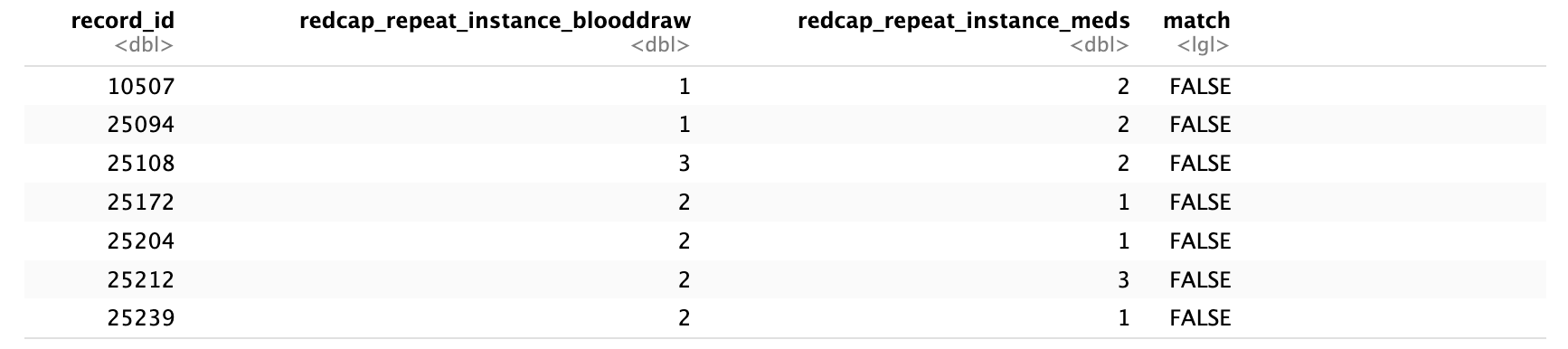
Thoracic

Proposed solution for data release 2.0: For the IDs above, we compared the time stamps of the Biospecimen collection Form CRF(s) and the Current Medications Form(s), and retained the Current Medications Form(s) with the time stamp matching or close to the Biospecimen collection Form CRF(s) dates.
bio <- bio %>%
filter(
!(record_id == 10507 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 2)
) %>%
filter(
!(record_id == 20326 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 2)
) %>%
mutate(
redcap_repeat_instance = case_when(
record_id == 25172 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 1 ~
2,
TRUE ~ redcap_repeat_instance
)
) %>%
mutate(
redcap_repeat_instance = case_when(
record_id == 25204 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 1 ~
2,
TRUE ~ redcap_repeat_instance
)
) %>%
filter(
!(record_id == 25212 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 1 |
redcap_repeat_instance == 3)
) %>%
mutate(
redcap_repeat_instance = case_when(
record_id == 25239 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 1 ~
2,
TRUE ~ redcap_repeat_instance
)
) %>%
filter(
!(record_id == 25094 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 1)
) %>%
mutate(
redcap_repeat_instance = case_when(
record_id == 25094 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 2 ~
1,
TRUE ~ redcap_repeat_instance
)
) %>%
mutate(
redcap_repeat_instance = case_when(
record_id == 25108 &
redcap_repeat_instrument == "current_medications_v02" &
redcap_repeat_instance == 2 ~
3,
TRUE ~ redcap_repeat_instance
)
)E.2.4 TKA cohort Biospecimen collection form
tka_bio <- bio %>%
select(
record_id,
guid,
redcap_data_access_group,
redcap_event_name,
redcap_repeat_instrument,
redcap_repeat_instance,
starts_with("bscp"),
blood_sample_collection_and_processing_crf_complete,
cohort
)keep subjects from the TKA cohort, with the most recent baseline visit.
tka_bio <- tka_bio %>%
filter(cohort == "TKA") %>%
filter(
redcap_repeat_instrument == "blood_sample_collection_and_processing_crf"
) %>%
filter(
blood_sample_collection_and_processing_crf_complete == 2 &
!is.na(redcap_repeat_instance)
) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))Remove subjects that haven’t come in for a visit yet i.e. No blood draw time available and ‘No blood obtained’ marked)
brdata1 <- tka_bio %>%
filter(
bscp_sample_obtained___1 == 0 &
blood_sample_collection_and_processing_crf_complete == 2
)E.2.4.1 Flag 1:
Check if there is missing info on hours since last drink.
brflag1 <- brdata1 %>%
filter(is.na(bscp_hrs_since_water)) %>%
add_column(error_type = "Missing info on hours since last drink") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.4.2 Flag 2:
Check if there is missing information on hours since last food.
brflag2 <- brdata1 %>%
filter(is.na(bscp_hrs_since_food)) %>%
add_column(error_type = "Missing information on hours since last food") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.4.3 Flag 3:
Check if there is missing information on hours since last caffeine intake in subjects who consume caffeine.
brflag3 <- brdata1 %>%
filter(is.na(bscp_hrs_since_cafstim) & bscp_caff_cups_amt != 4) %>%
add_column(
error_type = "Missing information on hours since last caffeine"
) %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.4.4 Flag 4:
Check if there is missing information the amount of caffeine.
brflag4 <- brdata1 %>%
filter(is.na(bscp_caff_cups_amt)) %>%
add_column(error_type = "Missing information on the amount of caffeine") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.4.5 Flag 5:
Check if there is missing information on vaccination.
brflag5 <- brdata1 %>%
filter(is.na(bscp_any_vacc)) %>%
add_column(error_type = "Missing information on vaccination") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.4.6 Create Biospecimen collection form error report for the TKA cohort.
# Specify the common prefix
br_error <- "brflag"
# Find data frames in the global environment with the specified prefix
br_list <- mget(ls(pattern = paste0("^", br_error)))
# Combine the data frames using bind_rows
br_report <- bind_rows(br_list) %>%
pivot_wider(names_from = "error_type", values_from = "errors") %>%
mutate_all(~ replace_na(., ""))br_report %>%
gt() %>%
tab_header(
title = md("**TKA Biospecimen collection form Error Report**")
) %>%
tab_options(
table.font.size = px(12),
column_labels.font.size = px(12)
) %>%
tab_style(
style = list(cell_fill(color = "#F4F4F4")),
locations = cells_body(columns = record_id)
)| TKA Biospecimen collection form Error Report | ||||||||
| record_id | redcap_data_access_group | redcap_repeat_instrument | redcap_repeat_instance | Missing info on hours since last drink | Missing information on hours since last food | Missing information on hours since last caffeine | Missing information on the amount of caffeine | Missing information on vaccination |
|---|---|---|---|---|---|---|---|---|
| 10066 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | error | ||
| 10083 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10098 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | error | ||
| 10170 | northshore | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 10202 | northshore | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 10329 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 10506 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10594 | northshore | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10618 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 10644 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 10703 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10775 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 25048 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 25061 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 25275 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10042 | northshore | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10055 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10239 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 25020 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10016 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10029 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10232 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10339 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10589 | uchicago | blood_sample_collection_and_processing_crf | 1 | error | error | |||
| 10038 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10127 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10562 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10565 | northshore | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10702 | rush_university_me | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10838 | northshore | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 10882 | northshore | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 25014 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 25055 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
E.2.4.7 Save:
Save “tka_bio” and data dictionary as .csv files in the folder named “Reformatted”
write_csv(
tka_bio,
file = here::here(
"data",
"blood-draw",
"Reformatted",
"reformatted_tka_bio.csv"
)
)
write_csv(
bio_dict,
file = here::here("data", "blood-draw", "Reformatted", "updated_bio_dict.csv")
)E.2.5 Thoracotomy Cohort Biospecimen Collection Form:
thor_bio <- bio %>%
select(
record_id,
guid,
redcap_data_access_group,
redcap_event_name,
redcap_repeat_instrument,
redcap_repeat_instance,
starts_with("bscp"),
blood_sample_collection_and_processing_crf_complete,
cohort
)keep subjects from the Thoracotomy cohort, with the most recent baseline visit.
thor_bio <- thor_bio %>%
filter(cohort == "Thoracic") %>%
filter(
redcap_repeat_instrument == "blood_sample_collection_and_processing_crf"
) %>%
filter(
blood_sample_collection_and_processing_crf_complete == 2 &
!is.na(redcap_repeat_instance)
) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))Remove subjects that haven’t come in for a visit yet i.e. No blood draw time available and ‘No blood obtained’ marked
tbrdata1 <- thor_bio %>%
filter(
bscp_sample_obtained___1 == 0 &
blood_sample_collection_and_processing_crf_complete == 2
)E.2.5.1 Flag 1:
Check if there is missing information on hours since last drink.
tbrflag1 <- tbrdata1 %>%
filter(is.na(bscp_hrs_since_water)) %>%
add_column(error_type = "Missing information on hours since last drink") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.5.2 Flag 2:
Check if there is missing information on hours since last food.
tbrflag2 <- tbrdata1 %>%
filter(is.na(bscp_hrs_since_food)) %>%
add_column(error_type = "Missing information on hours since last food") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.5.3 Flag 3:
Check if there is missing information on hours since last caffeine intake in subjects who consume caffeine.
tbrflag3 <- tbrdata1 %>%
filter(is.na(bscp_hrs_since_cafstim) & bscp_caff_cups_amt != 4) %>%
add_column(
error_type = "Missing information on hours since last caffeine"
) %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.5.4 Flag 4:
Check if there is missing information on the amount of caffeine.
tbrflag4 <- tbrdata1 %>%
filter(is.na(bscp_caff_cups_amt)) %>%
add_column(error_type = "Missing information on the amount of caffeine") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.5.5 Flag 5:
Check if there is missing information on vaccination.
tbrflag5 <- tbrdata1 %>%
filter(is.na(bscp_any_vacc)) %>%
add_column(error_type = "Missing information on vaccination") %>%
add_column(errors = "error") %>%
select(
record_id,
redcap_data_access_group,
redcap_repeat_instrument,
redcap_repeat_instance,
error_type,
errors
)E.2.5.6 Create Biospecimen collection form error report for the Thoracotomy cohort.
# Specify the common prefix
tbr_error <- "tbrflag"
# Find data frames in the global environment with the specified prefix
tbr_list <- mget(ls(pattern = paste0("^", tbr_error)))
# Combine the data frames using bind_rows
tbr_report <- bind_rows(tbr_list) %>%
pivot_wider(names_from = "error_type", values_from = "errors") %>%
mutate_all(~ replace_na(., ""))tbr_report %>%
gt() %>%
tab_header(
title = md(
"**Thoracotomy Cohort Biospecimen collection form Error Report**"
)
) %>%
tab_options(
table.font.size = px(12),
column_labels.font.size = px(12)
) %>%
tab_style(
style = list(cell_fill(color = "#F4F4F4")),
locations = cells_body(columns = record_id)
)| Thoracotomy Cohort Biospecimen collection form Error Report | ||||||||
| record_id | redcap_data_access_group | redcap_repeat_instrument | redcap_repeat_instance | Missing information on hours since last drink | Missing information on hours since last food | Missing information on hours since last caffeine | Missing information on the amount of caffeine | Missing information on vaccination |
|---|---|---|---|---|---|---|---|---|
| 20042 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20115 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20292 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | error | error | error | |
| 20013 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20061 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20081 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20117 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20125 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20240 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20078 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20121 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20189 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
| 20275 | university_of_mich | blood_sample_collection_and_processing_crf | 1 | error | ||||
E.2.5.7 Save:
Save “thor_bio” and data dictionary as .csv files in the folder named “Reformatted”
write_csv(
thor_bio,
file = here::here(
"data",
"blood-draw",
"Reformatted",
"reformatted_thor_bio.csv"
)
)
write_csv(
bio_dict,
file = here::here("data", "blood-draw", "Reformatted", "updated_bio_dict.csv")
)E.2.6 TKA cohort Current Medications v_02 form:
tka_meds <- bio %>%
select(
record_id,
guid,
redcap_data_access_group,
redcap_event_name,
redcap_repeat_instrument,
redcap_repeat_instance,
starts_with("cmc"),
current_medications_v02_complete,
cohort
)keep subjects from the TKA cohort, with the most recent baseline visit.
tka_meds <- tka_meds %>%
filter(cohort == "TKA") %>%
filter(redcap_repeat_instrument == "current_medications_v02") %>%
filter(
current_medications_v02_complete == 2 & !is.na(redcap_repeat_instance)
) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))E.2.6.1 Save:
Save “tka_meds” and data dictionary as .csv files in the folder named “Reformatted”
write_csv(
tka_meds,
file = here::here(
"data",
"blood-draw",
"Reformatted",
"reformatted_tka_meds.csv"
)
)E.2.7 Thoracotomy cohort Current Medications v_02 form:
thor_meds <- bio %>%
select(
record_id,
guid,
redcap_data_access_group,
redcap_event_name,
redcap_repeat_instrument,
redcap_repeat_instance,
starts_with("cmc"),
current_medications_v02_complete,
cohort
)keep subjects from the TKA cohort, with the most recent baseline visit.
thor_meds <- thor_meds %>%
filter(cohort == "Thoracic") %>%
filter(redcap_repeat_instrument == "current_medications_v02") %>%
filter(
current_medications_v02_complete == 2 & !is.na(redcap_repeat_instance)
) %>%
group_by(record_id, redcap_event_name) %>%
top_n(1, redcap_repeat_instance) %>%
ungroup() %>%
select(where(not_all_na))E.2.7.1 Save:
Save “thor_meds” and data dictionary as .csv files in the folder named “Reformatted”
write_csv(
thor_meds,
file = here::here(
"data",
"blood-draw",
"Reformatted",
"reformatted_thor_meds.csv"
)
)