/corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer5 FreeSurfer Measures
One of the core scans in the A2CPS neuroimaging protocol is a T1w scan, which is used to capture structural information. These scans are processed using a software suite called FreeSurfer. FreeSurfer takes the 3D image, segments it into pre-defined anatomical regions (like the hippocampus or amygdala), and also creates a triangular “mesh” of cerebral cortex from which it derives measurements of structural morphometry, including cortical thickness, surface area, and curvature. These measurements are related to a wide range of diseases, including chronic pain, and are often used as a source of features for predictive modeling. A2CPS has compiled the measurements into tabular files, providing a rich set of neuroanatomical variables ready for statistical analysis to explore brain-behavior relationships or group differences. This kit provides an overview of those tabular files.
5.1 Starting Project
5.1.1 Locate data
In the release, data are stored underneath the mris/derivatives folder:
This folder contains all of the FreeSurfer outputs for every participant (e.g., this is the directory that could correspond to the FreeSurfer environment variable $SUBJECTS_DIR), which subject folders of the form sub-[recordid]_ses-[protocolid].
Most users will not need the raw FreeSurfer outputs and can instead rely on tables in which the morphological measures have been aggregated. There are three relevant tables
- aparc.tsv
- measurements derived from parcellations of the cortical surface (e.g., cortical thickness)
- produced by
aparcstats2table
- aseg.tsv
- measurements derived from segmentations of the 3d structural image (e.g., average signal intensity)
- produced by
asegstats2table
- headers.tsv
- information about the brain as a whole (e.g., estimated Total Intracranial Volume)
- derived from the headers of the
aparcstats2tableandasegstats2tableoutputs
For each of these, there are data dictionaries in json files (e.g., aparc.json explains the columns of aparc.tsv).
$ ls /corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/*{json,tsv}
/corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/aparc.json /corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/gm_morph.tsv
/corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/aparc.tsv /corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/headers.json
/corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/aseg.json /corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/headers.tsv
/corral-secure/projects/A2CPS/products/consortium-data/pre-surgery/mris/derivatives/freesurfer/aseg.tsvCopies of the data dictionaries are available online for preview: aparc.json, aseg.json, headers.json.
5.1.2 Extract data
In this kit, we will explore the cortical parcellations.
library(readr)
library(dplyr)
library(tidyr)
library(ggplot2)As described above, the parcellation information is stored in aparc.tsv.
aparc <- read_tsv("data/aparc.tsv", show_col_types = FALSE)
head(aparc)| StructName | NumVert | SurfArea | GrayVol | ThickAvg | ThickStd | MeanCurv | GausCurv | FoldInd | CurvInd | sub | ses | hemisphere | parc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bankssts | 1360 | 909 | 2055 | 2.516 | 0.397 | 0.112 | 0.020 | 10 | 1.1 | 10706 | V1 | lh | aparc |
| caudalanteriorcingulate | 852 | 604 | 1626 | 2.253 | 0.887 | 0.131 | 0.024 | 13 | 1.0 | 10706 | V1 | lh | aparc |
| caudalmiddlefrontal | 2870 | 1857 | 4018 | 2.164 | 0.485 | 0.104 | 0.018 | 22 | 2.2 | 10706 | V1 | lh | aparc |
| cuneus | 1595 | 1090 | 2019 | 1.738 | 0.515 | 0.141 | 0.027 | 19 | 2.0 | 10706 | V1 | lh | aparc |
| entorhinal | 480 | 306 | 1366 | 3.029 | 0.816 | 0.105 | 0.018 | 4 | 0.3 | 10706 | V1 | lh | aparc |
| fusiform | 3434 | 2414 | 8326 | 2.907 | 0.603 | 0.123 | 0.023 | 38 | 3.4 | 10706 | V1 | lh | aparc |
That table contains nine regional measurements (GrayVol, SurfArea) for the various structures that compose each of six parcellations.
A typical analysis will only rely on one parcellation. For details of the different parcellations, see the FreeSurfer documentation. Two good starting choices are either aparc or aparc.a2009s. The aparc atlas is a coarser parcellation, whereas aparc.a2009s is finer. Here, we’ll filter for aparc.a2009s.
a2009s <- aparc |>
filter(parc == "aparc.a2009s")
head(a2009s)| StructName | NumVert | SurfArea | GrayVol | ThickAvg | ThickStd | MeanCurv | GausCurv | FoldInd | CurvInd | sub | ses | hemisphere | parc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G_and_S_frontomargin | 828 | 603 | 1341 | 2.098 | 0.476 | 0.131 | 0.026 | 10 | 0.9 | 10706 | V1 | lh | aparc.a2009s |
| G_and_S_occipital_inf | 1116 | 762 | 2218 | 2.474 | 0.643 | 0.127 | 0.025 | 11 | 1.1 | 10706 | V1 | lh | aparc.a2009s |
| G_and_S_paracentral | 1548 | 858 | 1810 | 1.886 | 0.580 | 0.097 | 0.019 | 10 | 1.5 | 10706 | V1 | lh | aparc.a2009s |
| G_and_S_subcentral | 1151 | 809 | 2142 | 2.443 | 0.413 | 0.129 | 0.021 | 13 | 1.0 | 10706 | V1 | lh | aparc.a2009s |
| G_and_S_transv_frontopol | 531 | 480 | 1424 | 2.378 | 0.585 | 0.203 | 0.052 | 11 | 1.3 | 10706 | V1 | lh | aparc.a2009s |
| G_and_S_cingul-Ant | 2005 | 1481 | 3804 | 2.463 | 0.531 | 0.140 | 0.028 | 31 | 2.5 | 10706 | V1 | lh | aparc.a2009s |
5.1.3 Example Analysis: Relationship between Cortical Surface Area and Gray Matter Volume
One of the most common features in predictive models is cortical thickness, but each of the morphological measurements may carry unique information. For example, cortical surface area may be more predictive of widespread chronic pain as compared to cortical thickness, which may be more closely related to chronic headaches (Bhatt et al., 2024). Let’s compare the regional volume with the regional surface area.
a2009s |>
ggplot(aes(x = SurfArea, y = GrayVol)) +
geom_point(alpha = 0.1)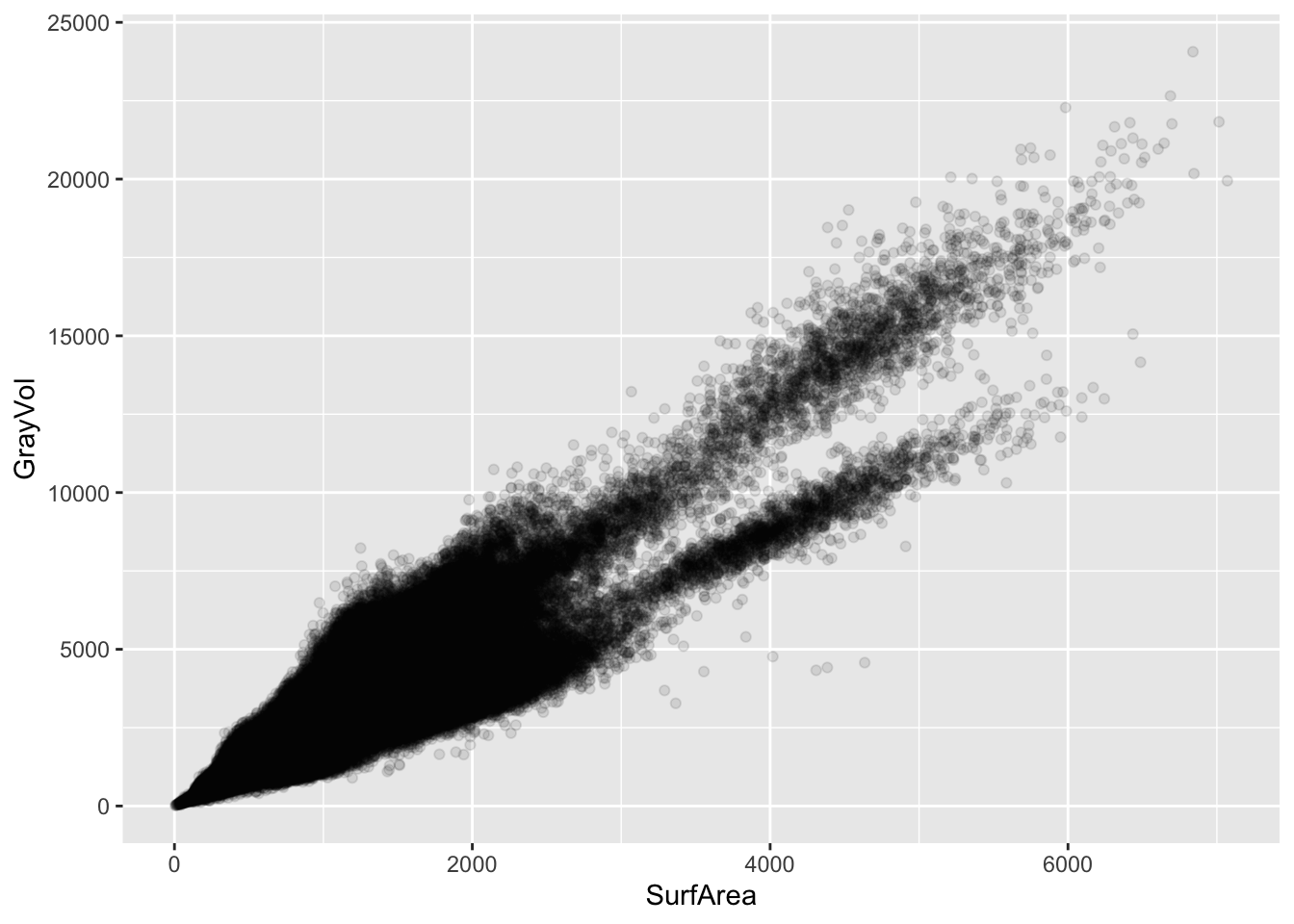
Clearly, there are groupings in the data. These groupings could partly be driven by participant demographics (e.g., some participants having larger or smaller regional volumes), but they may also be driven by different structures (e.g., the relationship between surface area and volume may differ across regions). Let’s zoom in on two of the larger clusters, and color the points by structure.
a2009s |>
filter(between(SurfArea, 4000, 6000), between(GrayVol, 10000, 20000)) |>
ggplot(aes(x = SurfArea, y = GrayVol, color = StructName)) +
geom_point()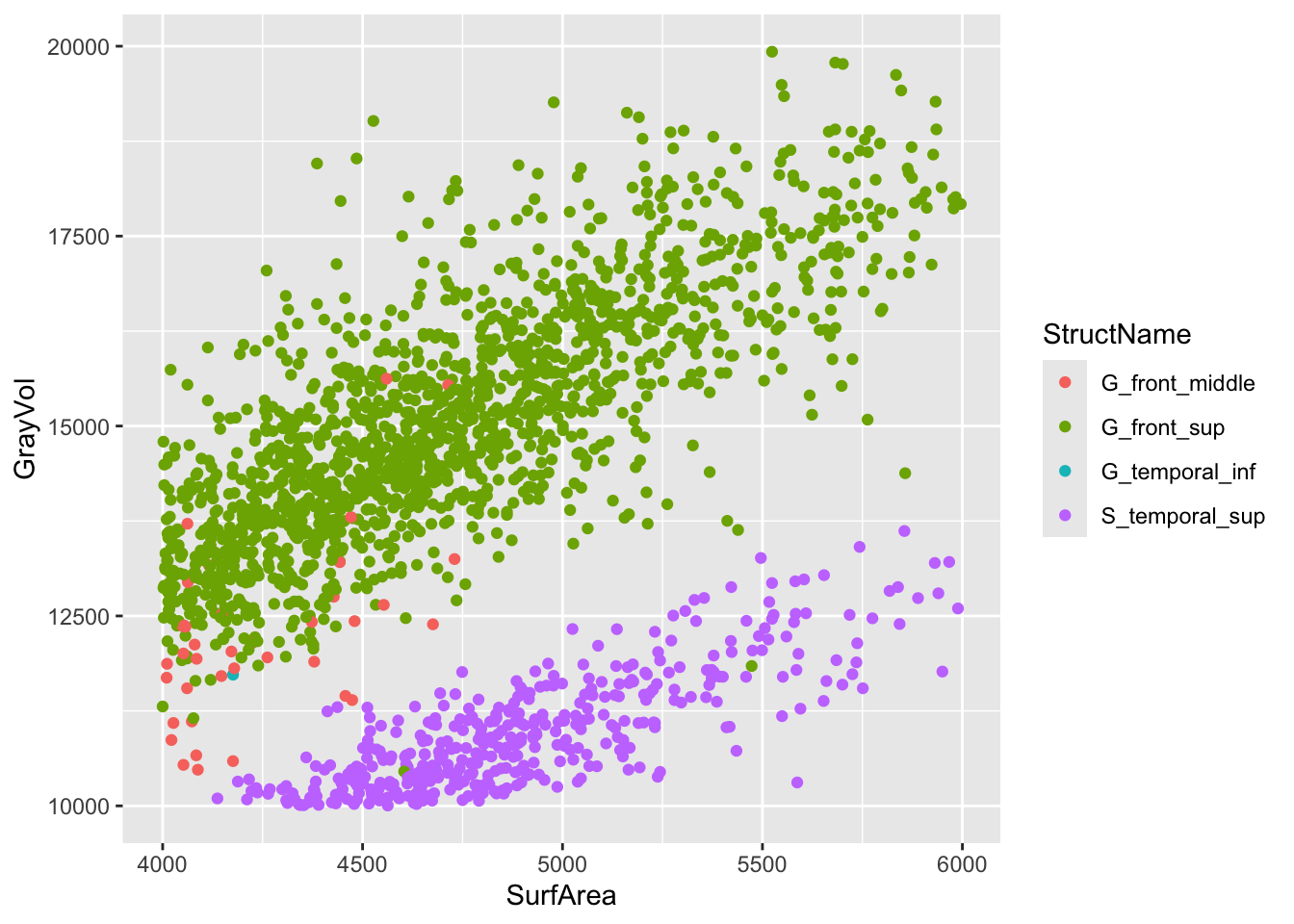
The relationship between surface area and volume appears to vary by region. For example, the Superior Temporal Sulcus has a similar surface area to the Superior Frontal Gyrus, but that gyrus has a larger volume.
5.1.4 Pivoting the Data
The data have been shared in a “longer” format, with rows corresponding to region-level observations. Some analyses benefit from having the data in a “wider” format, with rows corresponding to participant-level observations. For example, if we’re predicting age from cortical thickness, we may want to drop the other morphological measures, and pivot the table such that columns correspond to regional thickness.
StructName) has been combined with the hemisphere label.
a2009s |>
select(StructName, ThickAvg, sub, hemisphere) |>
pivot_wider(
names_from = c(StructName, hemisphere),
values_from = ThickAvg
) |>
head()| sub | G_and_S_frontomargin_lh | G_and_S_occipital_inf_lh | G_and_S_paracentral_lh | G_and_S_subcentral_lh | G_and_S_transv_frontopol_lh | G_and_S_cingul-Ant_lh | G_and_S_cingul-Mid-Ant_lh | G_and_S_cingul-Mid-Post_lh | G_cingul-Post-dorsal_lh | G_cingul-Post-ventral_lh | G_cuneus_lh | G_front_inf-Opercular_lh | G_front_inf-Orbital_lh | G_front_inf-Triangul_lh | G_front_middle_lh | G_front_sup_lh | G_Ins_lg_and_S_cent_ins_lh | G_insular_short_lh | G_occipital_middle_lh | G_occipital_sup_lh | G_oc-temp_lat-fusifor_lh | G_oc-temp_med-Lingual_lh | G_oc-temp_med-Parahip_lh | G_orbital_lh | G_pariet_inf-Angular_lh | G_pariet_inf-Supramar_lh | G_parietal_sup_lh | G_postcentral_lh | G_precentral_lh | G_precuneus_lh | G_rectus_lh | G_subcallosal_lh | G_temp_sup-G_T_transv_lh | G_temp_sup-Lateral_lh | G_temp_sup-Plan_polar_lh | G_temp_sup-Plan_tempo_lh | G_temporal_inf_lh | G_temporal_middle_lh | Lat_Fis-ant-Horizont_lh | Lat_Fis-ant-Vertical_lh | Lat_Fis-post_lh | Pole_occipital_lh | Pole_temporal_lh | S_calcarine_lh | S_central_lh | S_cingul-Marginalis_lh | S_circular_insula_ant_lh | S_circular_insula_inf_lh | S_circular_insula_sup_lh | S_collat_transv_ant_lh | S_collat_transv_post_lh | S_front_inf_lh | S_front_middle_lh | S_front_sup_lh | S_interm_prim-Jensen_lh | S_intrapariet_and_P_trans_lh | S_oc_middle_and_Lunatus_lh | S_oc_sup_and_transversal_lh | S_occipital_ant_lh | S_oc-temp_lat_lh | S_oc-temp_med_and_Lingual_lh | S_orbital_lateral_lh | S_orbital_med-olfact_lh | S_orbital-H_Shaped_lh | S_parieto_occipital_lh | S_pericallosal_lh | S_postcentral_lh | S_precentral-inf-part_lh | S_precentral-sup-part_lh | S_suborbital_lh | S_subparietal_lh | S_temporal_inf_lh | S_temporal_sup_lh | S_temporal_transverse_lh | G_and_S_frontomargin_rh | G_and_S_occipital_inf_rh | G_and_S_paracentral_rh | G_and_S_subcentral_rh | G_and_S_transv_frontopol_rh | G_and_S_cingul-Ant_rh | G_and_S_cingul-Mid-Ant_rh | G_and_S_cingul-Mid-Post_rh | G_cingul-Post-dorsal_rh | G_cingul-Post-ventral_rh | G_cuneus_rh | G_front_inf-Opercular_rh | G_front_inf-Orbital_rh | G_front_inf-Triangul_rh | G_front_middle_rh | G_front_sup_rh | G_Ins_lg_and_S_cent_ins_rh | G_insular_short_rh | G_occipital_middle_rh | G_occipital_sup_rh | G_oc-temp_lat-fusifor_rh | G_oc-temp_med-Lingual_rh | G_oc-temp_med-Parahip_rh | G_orbital_rh | G_pariet_inf-Angular_rh | G_pariet_inf-Supramar_rh | G_parietal_sup_rh | G_postcentral_rh | G_precentral_rh | G_precuneus_rh | G_rectus_rh | G_subcallosal_rh | G_temp_sup-G_T_transv_rh | G_temp_sup-Lateral_rh | G_temp_sup-Plan_polar_rh | G_temp_sup-Plan_tempo_rh | G_temporal_inf_rh | G_temporal_middle_rh | Lat_Fis-ant-Horizont_rh | Lat_Fis-ant-Vertical_rh | Lat_Fis-post_rh | Pole_occipital_rh | Pole_temporal_rh | S_calcarine_rh | S_central_rh | S_cingul-Marginalis_rh | S_circular_insula_ant_rh | S_circular_insula_inf_rh | S_circular_insula_sup_rh | S_collat_transv_ant_rh | S_collat_transv_post_rh | S_front_inf_rh | S_front_middle_rh | S_front_sup_rh | S_interm_prim-Jensen_rh | S_intrapariet_and_P_trans_rh | S_oc_middle_and_Lunatus_rh | S_oc_sup_and_transversal_rh | S_occipital_ant_rh | S_oc-temp_lat_rh | S_oc-temp_med_and_Lingual_rh | S_orbital_lateral_rh | S_orbital_med-olfact_rh | S_orbital-H_Shaped_rh | S_parieto_occipital_rh | S_pericallosal_rh | S_postcentral_rh | S_precentral-inf-part_rh | S_precentral-sup-part_rh | S_suborbital_rh | S_subparietal_rh | S_temporal_inf_rh | S_temporal_sup_rh | S_temporal_transverse_rh |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10706 | 2.098 | 2.474 | 1.886 | 2.443 | 2.378 | 2.463 | 2.445 | 2.534 | 2.532 | 2.524 | 1.669 | 2.481 | 2.420 | 2.099 | 2.239 | 2.400 | 2.852 | 3.408 | 2.156 | 2.121 | 2.971 | 1.903 | 2.697 | 2.380 | 2.142 | 2.550 | 1.817 | 1.848 | 2.371 | 2.196 | 2.159 | 2.267 | 2.179 | 2.897 | 3.292 | 2.471 | 2.957 | 2.683 | 1.688 | 1.982 | 2.160 | 1.851 | 3.500 | 1.601 | 1.646 | 1.756 | 2.585 | 2.545 | 2.344 | 2.892 | 1.939 | 2.058 | 1.990 | 2.073 | 1.959 | 1.878 | 1.843 | 1.698 | 2.021 | 2.702 | 2.294 | 1.998 | 2.186 | 2.444 | 1.947 | 1.429 | 2.086 | 2.166 | 2.099 | 2.098 | 2.134 | 2.398 | 2.359 | 2.337 | 2.316 | 2.112 | 1.715 | 2.466 | 2.542 | 2.462 | 2.429 | 2.348 | 2.722 | 2.145 | 1.736 | 2.361 | 2.381 | 2.222 | 2.153 | 2.392 | 3.494 | 3.292 | 2.324 | 1.759 | 2.829 | 1.966 | 2.858 | 2.613 | 2.285 | 2.430 | 1.827 | 1.740 | 2.227 | 2.035 | 2.136 | 2.505 | 2.048 | 2.859 | 3.178 | 2.231 | 2.638 | 2.809 | 1.878 | 1.938 | 2.346 | 1.942 | 3.461 | 1.479 | 1.630 | 1.940 | 2.644 | 2.709 | 2.369 | 3.315 | 2.103 | 1.880 | 2.097 | 1.956 | 2.364 | 1.841 | 1.870 | 1.840 | 2.208 | 2.126 | 2.256 | 1.793 | 2.458 | 2.636 | 1.884 | 1.368 | 1.765 | 2.217 | 2.097 | 2.145 | 2.283 | 2.536 | 2.348 | 2.297 |
| 10734 | 2.463 | 2.110 | 1.959 | 2.483 | 2.443 | 2.410 | 2.357 | 2.361 | 2.668 | 2.103 | 1.682 | 2.520 | 2.550 | 2.414 | 2.465 | 2.584 | 2.835 | 3.001 | 2.161 | 1.906 | 2.592 | 1.842 | 2.948 | 2.572 | 2.414 | 2.490 | 2.136 | 1.759 | 2.500 | 2.349 | 2.404 | 2.320 | 2.206 | 3.044 | 2.830 | 2.496 | 2.826 | 2.822 | 1.729 | 2.251 | 1.919 | 1.465 | 3.147 | 1.613 | 1.852 | 1.763 | 2.738 | 2.267 | 2.180 | 2.375 | 1.613 | 2.051 | 2.241 | 2.340 | 2.009 | 2.000 | 1.728 | 1.929 | 2.082 | 2.343 | 2.115 | 2.010 | 2.056 | 2.486 | 1.776 | 1.347 | 1.924 | 2.212 | 2.279 | 1.969 | 2.202 | 2.313 | 2.303 | 2.139 | 2.454 | 2.216 | 1.800 | 2.267 | 2.364 | 2.467 | 2.474 | 2.410 | 2.478 | 2.323 | 1.529 | 2.683 | 2.504 | 2.533 | 2.519 | 2.564 | 3.210 | 2.871 | 2.352 | 1.914 | 2.609 | 1.929 | 2.998 | 2.643 | 2.418 | 2.433 | 2.043 | 1.747 | 2.488 | 2.344 | 2.473 | 2.573 | 2.227 | 2.935 | 3.035 | 2.297 | 2.780 | 2.682 | 1.869 | 2.250 | 2.056 | 1.671 | 3.071 | 1.598 | 1.712 | 1.949 | 2.421 | 2.002 | 2.365 | 2.199 | 1.703 | 2.178 | 2.260 | 2.338 | 2.129 | 1.847 | 1.721 | 1.839 | 1.903 | 2.125 | 2.029 | 2.123 | 2.222 | 2.488 | 1.924 | 1.454 | 1.783 | 2.243 | 2.223 | 1.669 | 2.321 | 2.283 | 2.206 | 1.777 |
| 20371 | 2.498 | 2.080 | 2.218 | 2.804 | 2.722 | 2.690 | 2.394 | 2.328 | 2.520 | 2.367 | 1.757 | 2.570 | 2.602 | 2.463 | 2.496 | 2.611 | 3.370 | 3.746 | 2.028 | 1.820 | 2.795 | 1.917 | 2.864 | 2.680 | 2.608 | 2.666 | 2.199 | 1.985 | 2.593 | 2.470 | 2.379 | 2.572 | 2.651 | 3.030 | 3.124 | 2.709 | 2.750 | 3.101 | 1.714 | 2.197 | 2.380 | 1.694 | 3.276 | 1.723 | 1.885 | 2.165 | 2.564 | 2.951 | 2.371 | 2.657 | 2.008 | 2.022 | 2.088 | 2.183 | 2.705 | 2.094 | 1.664 | 1.848 | 2.032 | 2.591 | 2.380 | 2.208 | 2.165 | 2.689 | 2.165 | 1.553 | 2.117 | 2.262 | 2.161 | 2.611 | 2.225 | 2.652 | 2.461 | 2.404 | 2.405 | 2.615 | 2.079 | 2.522 | 2.431 | 2.537 | 2.252 | 2.325 | 2.645 | 2.415 | 1.583 | 2.650 | 2.496 | 2.285 | 2.403 | 2.605 | 3.346 | 3.601 | 2.128 | 1.915 | 2.648 | 1.912 | 2.948 | 2.548 | 2.671 | 2.509 | 2.293 | 1.955 | 2.397 | 2.416 | 2.668 | 2.713 | 2.523 | 2.940 | 3.145 | 2.521 | 2.736 | 2.956 | 2.127 | 2.363 | 2.290 | 1.621 | 3.067 | 1.695 | 1.788 | 2.314 | 2.900 | 2.658 | 2.485 | 2.501 | 1.979 | 2.128 | 2.161 | 2.139 | 2.051 | 2.150 | 1.652 | 2.002 | 2.242 | 2.441 | 2.276 | 2.087 | 2.212 | 2.540 | 2.155 | 1.335 | 2.132 | 2.353 | 2.179 | 2.147 | 2.135 | 2.507 | 2.489 | 2.502 |
| 20108 | 2.037 | 2.204 | 2.025 | 2.509 | 2.350 | 2.638 | 2.351 | 2.386 | 2.592 | 2.042 | 1.607 | 2.625 | 2.721 | 2.549 | 2.454 | 2.704 | 3.342 | 3.520 | 1.904 | 1.731 | 2.637 | 1.804 | 2.815 | 2.535 | 2.381 | 2.566 | 1.986 | 1.738 | 2.509 | 2.374 | 2.408 | 2.677 | 2.555 | 2.787 | 2.887 | 2.453 | 2.550 | 2.433 | 1.999 | 2.000 | 2.549 | 1.767 | 3.484 | 1.893 | 1.873 | 2.146 | 2.863 | 2.445 | 2.332 | 2.570 | 2.145 | 2.216 | 2.242 | 2.414 | 1.928 | 2.063 | 1.641 | 2.268 | 2.104 | 2.524 | 2.089 | 2.003 | 1.947 | 2.569 | 1.977 | 1.539 | 2.015 | 2.376 | 2.340 | 2.829 | 2.352 | 2.260 | 2.291 | 1.988 | 2.100 | 1.873 | 2.009 | 2.578 | 2.419 | 2.631 | 2.449 | 2.452 | 2.666 | 2.108 | 1.584 | 2.702 | 2.392 | 2.554 | 2.449 | 2.644 | 3.491 | 3.654 | 1.678 | 1.540 | 2.863 | 1.947 | 3.140 | 2.701 | 1.812 | 2.475 | 1.737 | 1.845 | 2.602 | 2.420 | 2.782 | 2.690 | 2.868 | 3.179 | 3.027 | 2.526 | 2.646 | 2.685 | 2.182 | 2.197 | 2.270 | 1.517 | 3.187 | 1.835 | 1.958 | 2.314 | 2.580 | 2.332 | 2.350 | 2.952 | 2.236 | 2.142 | 2.097 | 2.243 | 2.177 | 1.915 | 2.010 | 2.018 | 2.018 | 2.454 | 2.156 | 1.851 | 2.276 | 2.679 | 2.045 | 1.294 | 2.016 | 2.156 | 2.104 | 2.174 | 2.294 | 2.295 | 2.384 | 2.609 |
| 10689 | 2.345 | 2.583 | 2.608 | 2.687 | 2.610 | 2.623 | 2.684 | 2.545 | 2.691 | 2.234 | 1.962 | 2.591 | 2.724 | 2.583 | 2.717 | 2.899 | 3.240 | 3.609 | 2.442 | 1.845 | 2.933 | 1.905 | 3.182 | 2.940 | 2.673 | 2.870 | 2.453 | 2.343 | 2.955 | 2.615 | 2.394 | 2.531 | 2.287 | 3.181 | 3.027 | 2.703 | 3.118 | 3.226 | 2.061 | 2.427 | 2.344 | 1.840 | 3.380 | 1.668 | 2.004 | 2.186 | 2.830 | 2.620 | 2.625 | 2.468 | 2.123 | 2.352 | 2.258 | 2.474 | 2.269 | 2.283 | 1.823 | 2.255 | 2.312 | 2.395 | 2.624 | 2.292 | 2.224 | 2.531 | 2.327 | 1.340 | 2.321 | 2.452 | 2.378 | 2.271 | 2.500 | 2.506 | 2.473 | 2.629 | 2.515 | 2.916 | 2.410 | 2.480 | 2.688 | 2.589 | 2.584 | 2.578 | 2.885 | 2.423 | 1.789 | 2.744 | 2.604 | 2.590 | 2.674 | 2.959 | 3.512 | 3.546 | 2.732 | 2.115 | 2.735 | 2.166 | 3.210 | 2.994 | 2.656 | 2.857 | 2.509 | 2.286 | 2.874 | 2.617 | 2.564 | 2.791 | 2.548 | 3.309 | 3.292 | 2.520 | 2.986 | 3.309 | 2.311 | 2.370 | 2.452 | 1.833 | 3.422 | 1.892 | 2.015 | 2.269 | 2.597 | 2.487 | 2.745 | 2.415 | 2.194 | 2.247 | 2.414 | 2.610 | 2.199 | 2.221 | 2.089 | 2.210 | 2.323 | 2.408 | 2.473 | 2.211 | 2.229 | 2.627 | 2.333 | 1.701 | 2.173 | 2.507 | 2.394 | 2.447 | 2.542 | 2.513 | 2.538 | 2.878 |
| 20020 | 2.174 | 1.939 | 2.221 | 2.647 | 2.285 | 2.480 | 2.376 | 2.230 | 2.691 | 2.316 | 1.839 | 2.571 | 2.639 | 2.492 | 2.392 | 2.645 | 3.024 | 3.446 | 2.271 | 1.817 | 2.633 | 1.922 | 3.039 | 2.719 | 2.542 | 2.691 | 2.331 | 2.113 | 2.698 | 2.556 | 2.578 | 2.367 | 2.335 | 2.973 | 3.308 | 2.368 | 2.857 | 3.071 | 2.407 | 2.082 | 2.283 | 1.696 | 3.297 | 1.697 | 1.898 | 1.951 | 2.458 | 2.292 | 2.405 | 2.883 | 1.624 | 2.066 | 1.927 | 2.245 | 2.308 | 2.022 | 1.779 | 1.844 | 2.055 | 2.198 | 2.288 | 1.773 | 2.102 | 2.493 | 1.997 | 1.399 | 2.134 | 2.143 | 2.195 | 2.036 | 2.128 | 2.405 | 2.392 | 1.951 | 2.379 | 2.182 | 2.360 | 2.670 | 2.211 | 2.445 | 2.185 | 2.361 | 2.840 | 2.615 | 1.834 | 2.545 | 2.475 | 2.005 | 2.511 | 2.608 | 3.096 | 3.468 | 2.531 | 2.030 | 2.639 | 1.907 | 2.977 | 2.760 | 2.483 | 2.579 | 2.241 | 2.107 | 2.909 | 2.398 | 2.629 | 2.576 | 2.572 | 3.021 | 3.178 | 2.420 | 2.895 | 2.973 | 2.195 | 2.464 | 2.276 | 1.748 | 3.388 | 1.782 | 1.826 | 2.057 | 2.591 | 2.321 | 2.555 | 2.626 | 1.659 | 2.128 | 1.991 | 2.214 | 2.098 | 1.940 | 1.970 | 1.942 | 2.161 | 2.534 | 2.189 | 1.899 | 2.434 | 2.581 | 1.897 | 1.570 | 1.994 | 2.275 | 2.271 | 1.827 | 2.143 | 2.310 | 2.375 | 2.440 |
5.1.5 Variability Across Scanners
Many MRI biomarkers exhibit variability across the scanners, which may confound some analyses. For an up-to-date assessment of the issue and overview of current thinking, please see Confluence.
5.1.6 Data Quality
As with any MRI derivative, all pipeline derivatives have been included. This means that products were included regardless of their quality, and so some products may have been generated from images that are known to have poor quality—rated “red”, or incomparable. For details on the ratings and how to exclude them, see Appendix A. Additionally, extensive QC has not yet been performed on the derivatives themselves, and so there may be cases where pipelines produced atypical outputs. For an overview of planned checks, see Confluence.
Currently, no QC has been performed on the underlying segmentations and parcellations. With adequate sample sizes, results based on whole-atlas FreeSurfer measurements are generally robust, although there are individual regions that may warrant close inspection (e.g., McCarthy et al. 2015, Vahermaa et al. 2023). Moreover, it remains unclear the extent to which FreeSurfer’s accuracy persists over the wide range of demographics found in the A2CPS dataset. The Imaging DIRC is actively exploring these aspects of quality.
5.1.7 Methods
FreeSurfer outputs were generated by the fMRIPrep pipeline. The fMRIPrep pipeline is similar to a typical call to FreeSurfer’s recon-all, with a majority of changes aiming to parallelize more parts of recon-all. However, there are differences, such as in how the brain is masked. The A2CPS use of fMRIPrep is even more different because it entails replacing the fMRIPrep masking procedure with SynthStrip. In general, these changes are expected to only improve the quality of the FreeSurfer outputs. However, they may hinder some comparisons between results derived from A2CPS and those in other studies.
5.1.8 Citations
The FreeSurfer package has been developed through extensive research. If you use these derivatives in your analyses, please follow the documentation for citing FreeSurfer: https://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurferMethodsCitation.
In publications or presentations including data from A2CPS, please include the following statement as attribution:
Data were provided [in part] by the A2CPS Consortium funded by the National Institutes of Health (NIH) Common Fund, which is managed by the Office of the Director (OD)/ Office of Strategic Coordination (OSC). Consortium components and their associated funding sources include Clinical Coordinating Center (U24NS112873), Data Integration and Resource Center (U54DA049110), Omics Data Generation Centers (U54DA049116, U54DA049115, U54DA049113), Multi-site Clinical Center 1 (MCC1) (UM1NS112874), and Multi-site Clinical Center 2 (MCC2) (UM1NS118922).
When using neuroimaging derivatives, please also cite Sadil et al. (2024).